## 'data.frame': 4090 obs. of 29 variables:
## $ Production.Year : int 2019 2019 2019 2019 2017 2019 2018 2018 2017 2019 ...
## $ Production.Month : int 2 2 2 2 2 7 3 2 3 3 ...
## $ Product.Line : chr "Premium Pot" "Premium End Cap" "Premium Pot" "Basic End Cap" ...
## $ Genus : chr "Ipomoea" "Begonia" "Ipomoea" "Sunpatiens" ...
## $ SalesPlanQty : int 2500 2000 2500 2000 2000 1500 0 0 500 2000 ...
## $ LYSalesPlanQty : int 2500 2500 2500 2500 2500 2500 2500 2500 2500 2500 ...
## $ L2YSalesPlanQty : int 2500 2000 2500 2000 2500 1500 0 0 0 2000 ...
## $ CustomerComp : num NA -0.9556 NA -0.0529 -0.0226 ...
## $ GenusComp : num -0.0487 -0.0563 -0.0487 -0.057 -0.057 ...
## $ ProductLineComp : num -0.0609 -0.0659 -0.0609 -0.0126 -0.0431 ...
## $ CustomerMarginPerc : num 0.263 -0.198 0.416 0.35 0.383 ...
## $ CustomerMarginPercAllYear: num 0.2629 0.0953 0.416 0.3263 0.3884 ...
## $ CustomerST : num NA NA NA 0.843 1.187 ...
## $ CustomerSTAllYear : num NA NA NA 0.915 0.911 ...
## $ Customer.POS.Qty : int 8 20 7 1832 1111 2273 19 416 137 191 ...
## $ Shipped.Qty : int 0 0 0 2172 936 1638 324 1440 1092 594 ...
## $ Loss.. : num NA -1 NA -0.00429 -0.0017 ...
## $ PriorMonthPOS : int 0 162 0 1516 3004 4544 -7 154 -25 0 ...
## $ Prior2MonthPOS : int 0 0 0 0 0 5432 -6 0 -7 0 ...
## $ Prior3MonthPOS : int 0 0 0 0 0 2402 0 0 0 0 ...
## $ PostMonthPOS : int 0 0 0 2092 1269 484 734 2838 1512 48 ...
## $ Post2MonthPOS : int 0 0 0 3220 2484 31 2518 8249 2256 1 ...
## $ Post3MonthPOS : int 0 0 0 3580 2095 -2 1618 18134 3719 -3 ...
## $ PriorMonthShipped : int 0 0 0 984 1644 2322 0 420 0 0 ...
## $ Prior2MonthShipped : int 0 0 0 0 0 5742 0 0 0 0 ...
## $ Prior3MonthShipped : int 0 0 0 0 0 5310 0 0 0 0 ...
## $ PostMonthShipped : int 0 0 0 3336 2448 0 1860 4200 1476 0 ...
## $ Post2MonthShipped : int 0 0 0 3036 2904 0 2964 18190 3576 0 ...
## $ Post3MonthShipped : int 0 0 0 3708 2448 0 708 18950 3456 0 ...
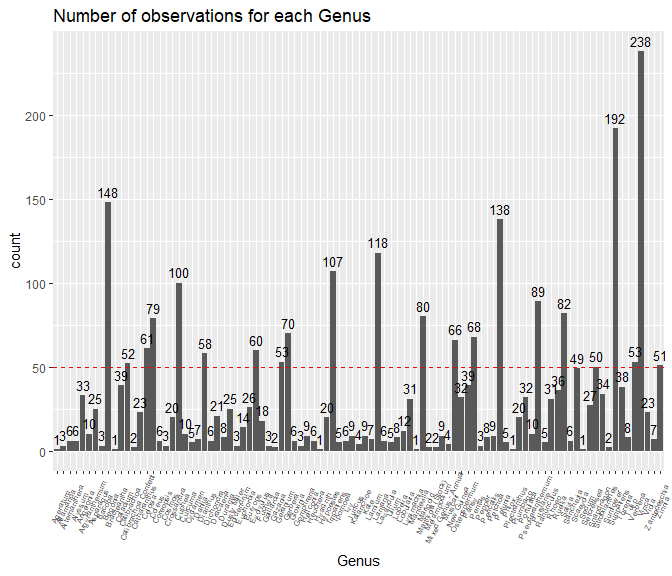
sales_data%>%
dplyr::group_by(Genus)%>%
dplyr::summarize(count =length(Genus))%>%
ggplot(., aes(x = Genus, y = count)) +
geom_bar(stat = "identity") +
ggtitle("Number of observations for each Genus")+
geom_text(aes(label=count), vjust=-.5, size = 3.5)+
theme(axis.text.x = element_text(angle=65, vjust=0.6, size = 6))+
geom_abline(slope=0,intercept=50, col= "red" , lty=2)
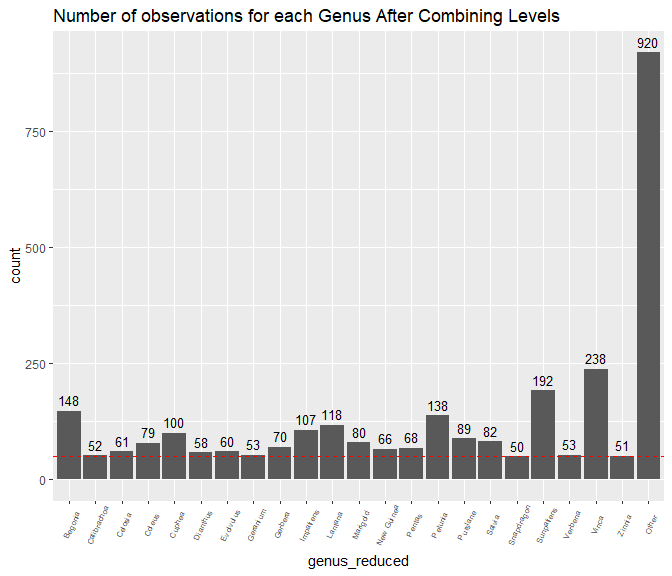
sales_data%>%
dplyr::group_by(genus_reduced)%>%
dplyr::summarize(count =length(genus_reduced))%>%
ggplot(., aes(x = genus_reduced, y = count)) +
geom_bar(stat = "identity") +
ggtitle("Number of observations for each Genus After Combining Levels")+
geom_text(aes(label=count), vjust=-.5, size = 3.5)+
theme(axis.text.x = element_text(angle=65, vjust=0.6, size = 6))+
geom_abline(slope=0,intercept=50, col= "red" , lty=2)
factor_vars <- names(sales_data)[which(sapply(sales_data,is.factor))]
dummies <- caret::dummyVars(~.,sales_data[factor_vars])
hot_coded <- stats::predict(dummies,sales_data[factor_vars])
numeric_vars <- names(sales_data)[which(sapply(sales_data,is.numeric))]
num_df <- sales_data[numeric_vars]
sales_data_onehot <- cbind(num_df,hot_coded)
trainSet <- sales_data_onehot[ which(sales_data_onehot[['Production.Year.2019']]=='0'),]
testSet <- sales_data_onehot[ which(sales_data_onehot[['Production.Year.2019']]=='1'),]
trainSet <- trainSet%>%
select(-c(Production.Year.2017,Production.Year.2018,Production.Year.2019,ID))
testSet <- testSet%>%
select(-c(Production.Year.2017,Production.Year.2018,Production.Year.2019,ID))
TARGET.VAR <- "SalesPlanQty"
candidate.features <- setdiff(names(trainSet),c(TARGET.VAR))
control <- trainControl(method="cv",
number=5,
allowParallel = TRUE,
savePredictions = 'final',
index=createFolds(y = trainSet$SalesPlanQty, 5))
algorithmList <- c('xgbTree','gbm','treebag','rf','xgbLinear', 'glm', 'knn', 'svmRadial')
set.seed(123)
output <- capture.output(
models <- caretList(trainSet[,candidate.features],
trainSet[,TARGET.VAR],
trControl=control,
methodList=algorithmList)
)
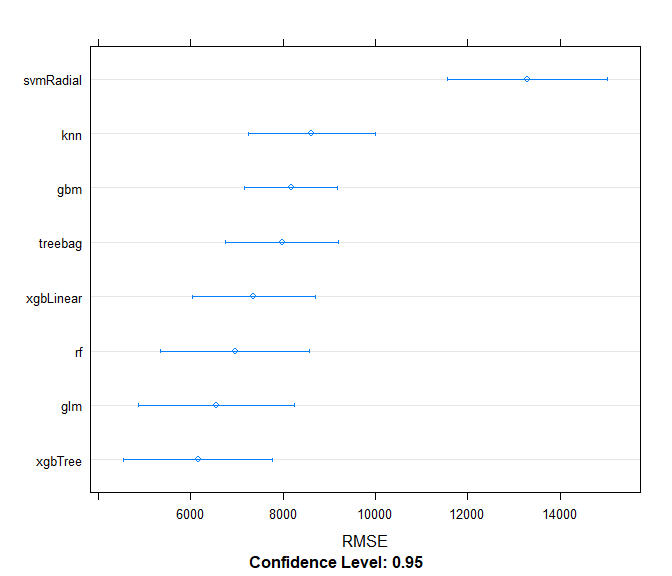
results <- resamples(models)
summary(results)
##
## Call:
## summary.resamples(object = results)
##
## Models: xgbTree, gbm, treebag, rf, xgbLinear, glm, knn, svmRadial
## Number of resamples: 5
##
## MAE
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## xgbTree 2637.428 2721.985 2726.128 2776.790 2743.828 3054.581 0
## gbm 3032.203 3043.684 3111.276 3199.624 3210.856 3600.104 0
## treebag 3099.639 3119.745 3176.998 3179.753 3227.054 3275.329 0
## rf 2437.218 2446.596 2661.226 2637.262 2819.315 2821.954 0
## xgbLinear 2644.383 2696.732 2748.980 2813.607 2882.822 3095.119 0
## glm 3037.855 3081.852 3097.342 3169.864 3118.863 3513.408 0
## knn 3127.012 3158.533 3296.377 3266.376 3332.952 3417.007 0
## svmRadial 3977.914 4176.274 4359.668 5173.783 4613.576 8741.483 0
##
## RMSE
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## xgbTree 5477.191 5554.174 5664.399 6382.935 6277.512 8941.399 0
## gbm 7615.189 7750.458 8211.933 8465.618 8874.726 9875.786 0
## treebag 6584.800 6924.217 7136.529 7723.727 8718.669 9254.422 0
## rf 5377.780 5560.336 6512.267 6954.287 8442.492 8878.560 0
## xgbLinear 5584.139 6058.581 7062.286 7004.103 7742.407 8573.100 0
## glm 5574.834 5839.821 6459.485 6304.812 6621.339 7028.579 0
## knn 7904.087 7920.332 8206.587 8802.588 9591.169 10390.765 0
## svmRadial 11305.672 11875.741 12299.283 14097.678 13773.012 21234.683 0
##
## Rsquared
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## xgbTree 0.8289601 0.9052816 0.9117711 0.8953623 0.9123067 0.9184920 0
## gbm 0.7958108 0.8138516 0.8262172 0.8214738 0.8291789 0.8423105 0
## treebag 0.8197593 0.8269932 0.8547851 0.8491072 0.8631677 0.8808310 0
## rf 0.8353103 0.8355050 0.8795324 0.8772529 0.9150776 0.9208391 0
## xgbLinear 0.8389560 0.8583360 0.8820060 0.8795809 0.9022909 0.9163156 0
## glm 0.8672814 0.8877565 0.9088949 0.8980954 0.9116003 0.9149439 0
## knn 0.7972584 0.8082220 0.8366949 0.8261298 0.8381716 0.8503024 0
## svmRadial 0.5585796 0.6055735 0.6402596 0.6304875 0.6651735 0.6828510 1
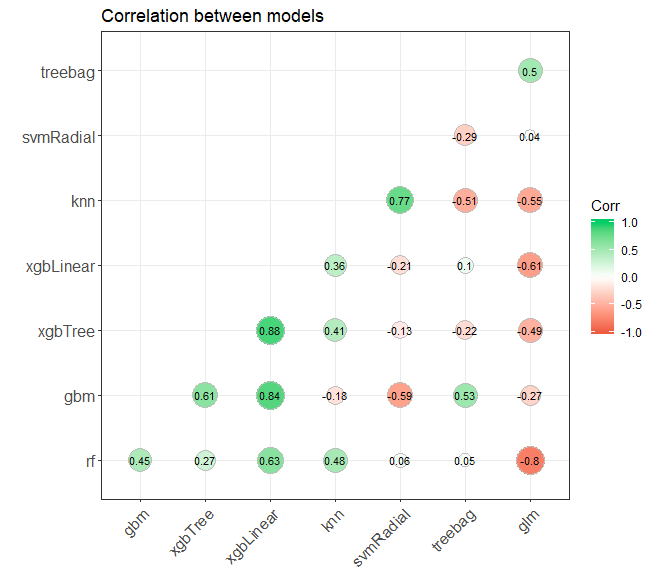
modelcorr <- modelCor(results)
ggcorrplot(modelcorr, hc.order = TRUE,
type = "lower",
lab = TRUE,
lab_size = 3,
method="circle",
colors = c("tomato2", "white", "springgreen3"),
title="Correlation between models",
ggtheme=theme_bw)
preds$pred_xgbtree <- predict.train(object = models$xgbTree, newdata = testSet[,candidate.features])
preds$pred_gbm <- predict.train(object = models$gbm, newdata = testSet[,candidate.features])
preds$pred_treebag <- predict.train(object = models$treebag, newdata = testSet[,candidate.features])
preds$pred_rf <- predict.train(object = models$rf, newdata = testSet[,candidate.features])
preds$pred_xgblin <- predict.train(object = models$xgbLinear, newdata = testSet[,candidate.features])
preds$pred_glm <- predict.train(object = models$glm, newdata = testSet[,candidate.features])
preds$pred_knn <- predict.train(object = models$knn, newdata = testSet[,candidate.features])
preds$pred_svmRadial <- predict.train(object = models$svmRadial, newdata = testSet[,candidate.features])
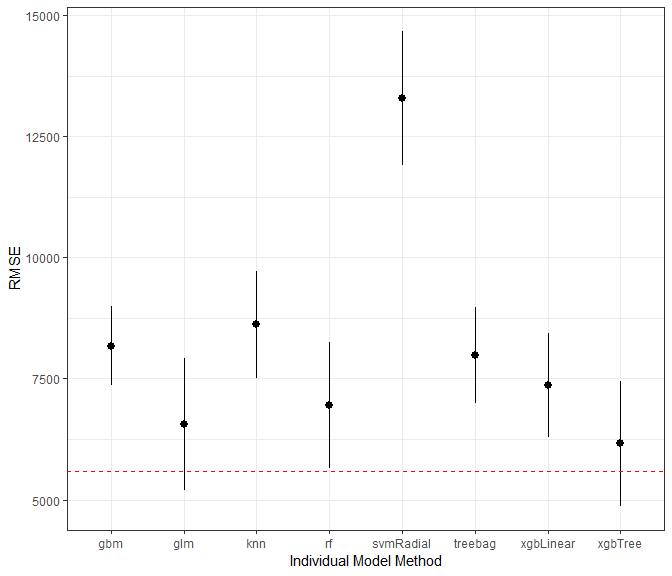
test_results <- data.frame(rmse = rapply(preds[,(ncol(preds)-7):ncol(preds)], function(x)RMSE(x,testSet[,TARGET.VAR])))
test_results$rsquared <- rapply(preds[,(ncol(preds)-7):ncol(preds)], function(x)R2(x,testSet[,TARGET.VAR]))
test_results$mae <- rapply(preds[,(ncol(preds)-7):ncol(preds)], function(x)MAE(x,testSet[,TARGET.VAR]))
preds$ensemble_1 <-predict(object = ensemble_1,newdata = testSet[,candidate.features])
preds$ensemble_2 <-predict(object = ensemble_2,newdata = testSet[,candidate.features])
#Below are the results of each of the models on the test dataset.
test_results <- data.frame(rmse = rapply(preds[,(ncol(preds)-9):ncol(preds)], function(x)RMSE(x,testSet[,TARGET.VAR])))
test_results$rsquared <- rapply(preds[,(ncol(preds)-9):ncol(preds)], function(x)R2(x,testSet[,TARGET.VAR]))
test_results$mae <- rapply(preds[,(ncol(preds)-9):ncol(preds)], function(x)MAE(x,testSet[,TARGET.VAR]))
kable(test_results)%>%
kable_styling(bootstrap_options = c("striped", "hover"), full_width = F)